Group leader: Laurence Delhaes
Group members: Raphaël Enaud, Sébastien Imbert, Gauthier Chauvin, Baptiste Defaye
As other mucosal sites of human body, respiratory tract hosts a specific microbiome leading to mutually beneficial interactions between humans and their microbiome (Jaggi et al. 2024; Chiu et al. 2017). The lung microbiome, which is believed to be stable or at least transient in healthy people, is known to be a poly-microorganism flora contributing to disease pathogenesis of both chronic pulmonary diseases such as chronic obstructive pulmonary disease (COPD), severe asthma and cystic fibrosis (CF) or acute infections, such as viral pneumonia or ventilator-acquired pneumonia. In turn, such pulmonary diseases can modify the lung microbiome composition leading to an increased risk of emerging or secondary pulmonary infections. Furthermore, respiratory tract is one of the major routes for emerging pathogens and infectious diseases (EID). Most research studies on lung microbiome have focused on bacteria and their impact on lung health, but there is evidence that other nonbacterial organisms, constituting the virome or mycobiome, are playing an important physio-pathological role.
In the last few years, our research group focused on lung mycobiome and microbiome analysis (Imbert et al. 2024; Enaud et al. 2023; Prevel et al. 2022; Delhaes et al. 2018; Nguyen et al. 2016) in relation with COPD, asthma, CF and EID, and its links with microbial exposome (Vandenborght et al. 2021).
Current project
Our major aim is to decipher the relationship between fungal and bacterial populations of lungs, inflammation, and other covariates such disease subtypes, indoor environment (exposome), or EID by combining (i) mycobiome-microbiome analysis, (ii) computational inter-kingdom and/or inter-organ network analysis (especially analysis of the gut-lung axis), (iii) ecological model investigation, (iv) microbial exposome analysis, and (v) in-vitro and/or in-vivo experiments, and (vi) by developing new investigation tools.
Collectively, these approaches allowed us to highlight the complexity of all respiratory microbiome components’ connections, which might yield important insights within chronic pulmonary diseases and EID.
Main objectives
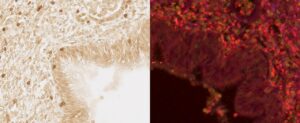
1. To assess lung mycobiome evolution during asthma, COPD and CF, especially during exacerbation
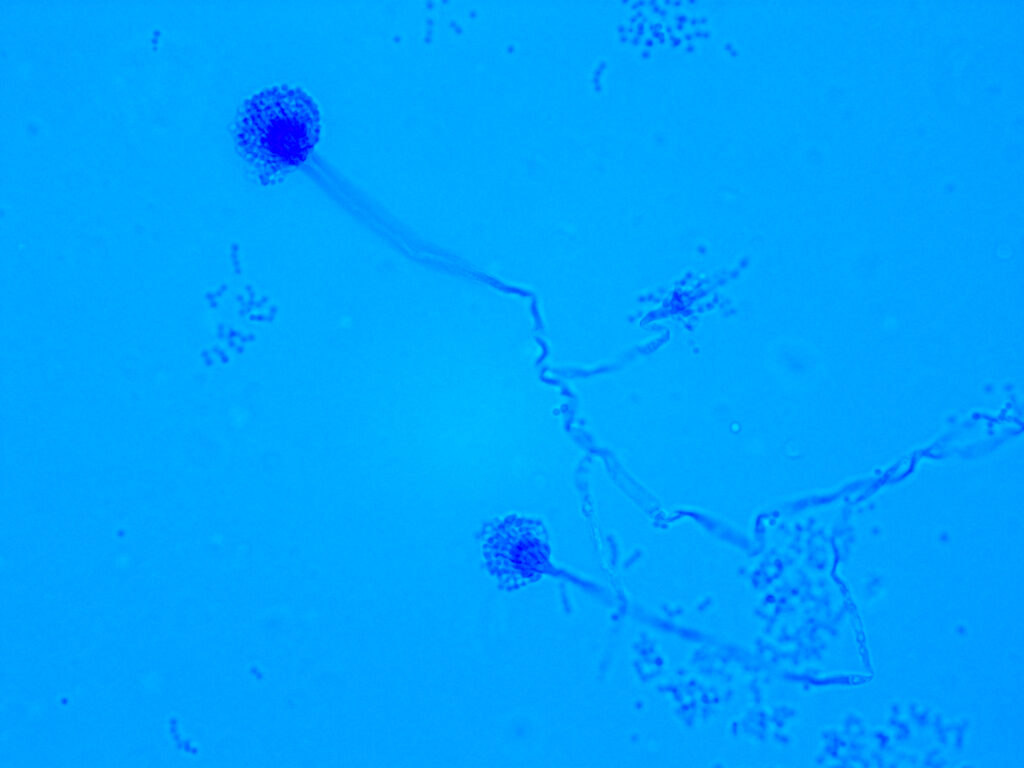
2. To decipher in these diseases, specific pulmonary fungal complications, such as severe asthma with fungal sensitization or secondary aspergillosis, and how fungal component will interact with microbial flora and/or host components
3. To investigate the clinical relevance of the “gut-lung axis” in chronic pulmonary diseases and/or EID
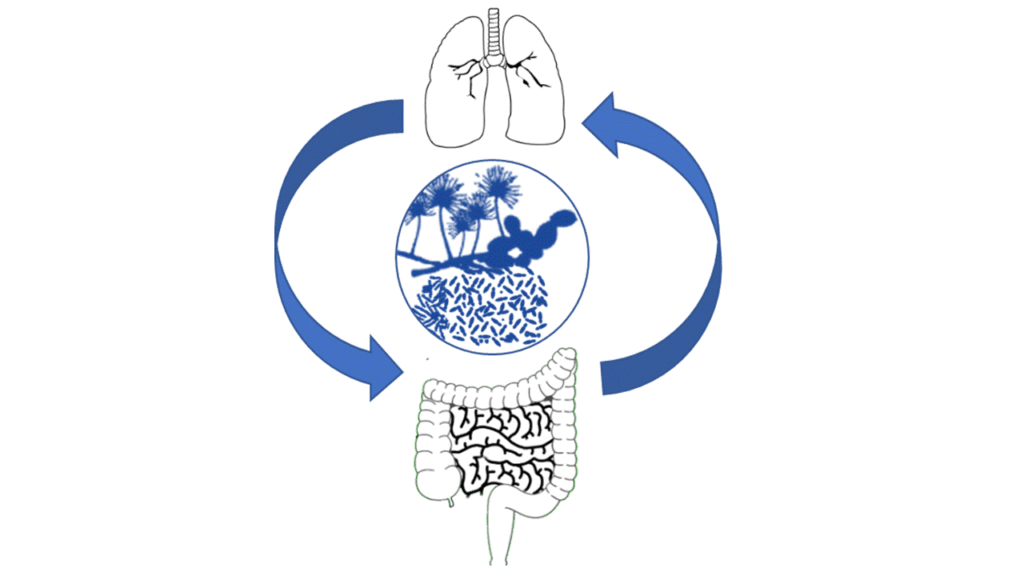
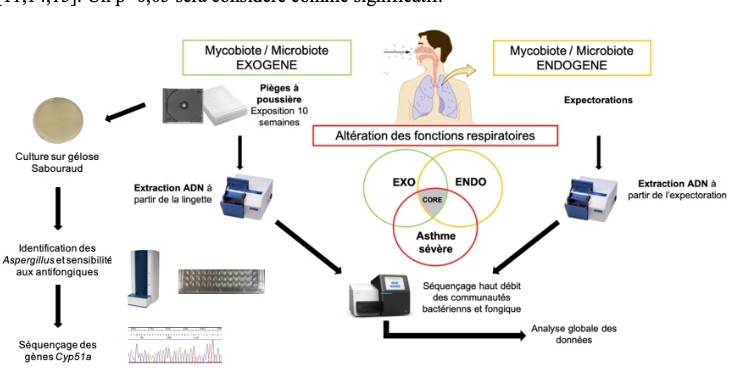
4. To study the role of exogenous mycobiome (fungal exposome) in these diseases
Main fundings: ANR « Inf-HOLOBIONT », PSGAR-MIE « EMERG », VLM association grant, Research grants of Bordeaux University, APITHEM GIRCI SOHO “MyCADO”, research grant from Société de Réanimation de Langue Française.
Our research fields are also linked to the CNR-AspC (National Reference Center of chronic aspergillosis-South region) coordinated by Pr L Delhaes at Bordeaux’s CHU – https://catbio.chu-bordeaux.fr/prog/defindex.php?idCor=Cor_bf3f2e66996a710964bb86fa77b95446.
References
- Imbert S, Revers M, Enaud R, Orieux A, Camino A, Massri A, Villeneuve L, Carrié C, Petit L, Boyer A, Berger P, Gruson D, Delhaes L, Prével R. Lower airway microbiota compositions differ between influenza, COVID-19 and bacteria-related acute respiratory distress syndromes. Crit Care. 2024 Apr 22;28(1):133.
- Jaggi TK, Agarwal R, Tiew PY, Shah A, Lydon EC, Hage CA, Waterer GW, Langelier CR, Delhaes L, Chotirmall SH. Fungal lung disease. Eur Respir J. 2024;64(5):2400803.
- F Lussac-Sorton, É Charpentier, S Imbert, M Lefranc, S Bui, M Fayon, P. Berger, R Enaud, L Delhaes. The gut-lung axis in the CFTR modulator era. Front. cell. infect. microbiol. 2023.
- Enaud R, Sioniac P, Imbert S, Janvier PL, Camino A, Bui HN, Pillet O, Orieux A, Boyer A, Berger P, Gruson D, Delhaes, Pr̩ével R. Lung mycobiota α-diversity is linked to severity in critically ill patients with acute exacerbation of chronic obstructive pulmonary disease. Microbiology Spectrum. 2023 Mar 28:e0506222.
- Prevel R, Enaud R, Orieux A, Camino A, Berger P, Boyer A, Delhaes L, Gruson D. Gut bacteriobiota and mycobiota can both predict day-28 mortality among critically ill patients. Crit care. 2022. 26(1):105.
- Prattes J, Wauters J, Giacobbe DR, Lagrou K, Hoenigl M, ECMM-CAPA Study Group. Diagnosis and treatment of COVID-19 associated pulmonary apergillosis in critically ill patients: results from a European confederation of medical mycology registry. Intensive Care Med 2021; 47: 1158-1160
- LE Vandenborght, R Enaud, C Urien, N Coron, P-O Girodet, S Ferreira, P Berger, L Delhaes. Type 2–high asthma is associated with a specific indoor mycobiome and microbiome. J Allergy Clin Immunol. 2020; 147: 1296-1305.e6.